Cancer
Leveraging Cross-Modality Interactions of DNA Methylation and Gene Expression for Insights Into Tumorigenesis in Lung Adenocarcinoma Shaira Kee* Shaira Kee Michael Aaron Sy Panayiotis V. Benos
Lung adenocarcinoma (LUAD) is the most common lung cancer in the United States. While treatment has improved, LUAD remains the leading cause of cancer death in the US. Studies show that conventional staging alone is insufficient in predicting LUAD prognosis and guiding treatment. Gene expression (GE) is among the instrumental tools in cancer research. Additionally, Aberrant DNA methylation (DNAm) is considered a hallmark of tumorigenesis and is associated with multiple malignancies.
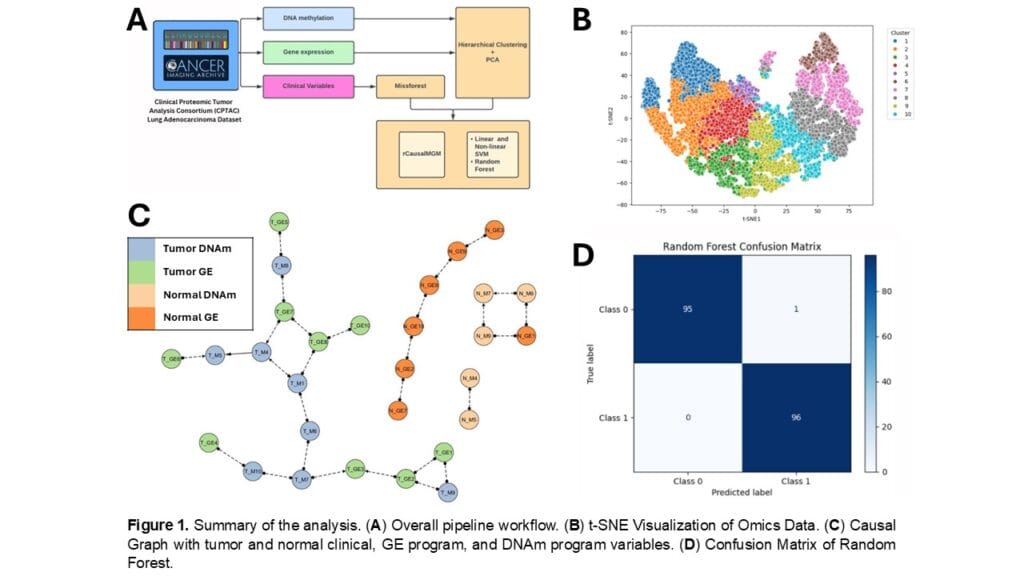
Clinical Proteomic Tumor Analysis Consortium (CPTAC) is a cancer research initiative that utilizes molecular data and matched pairs of tumor and normal tissue extracted from the same patient, which better assesses inter-patient variability and tumor-specific alterations. RNA-seq data from 96 CPTAC-LUAD participants were used in the study. Firstly, we extracted 10 distinct groups from the RNA-seq data (n = 15,314 genes) using ward linkage hierarchical clustering to reduce complexity. Subsequently, cross-modality interactions were analyzed using causal graph modeling of combined normalized clinical, GE, and DNAm.
We found a distinct separation of tumor and non-tumor omics data using causal graphs (see Fig. 1C), suggesting that certain gene groups are more indicative of tumor characteristics. The predictive classification using a random forest classifier achieved high performance with an accuracy of 99%, underscoring its robustness in differentiating tumors from normal samples.
In summary, this study proposes a new approach to determining gene signatures relevant to LUAD prognosis using causal graph approaches and machine learning prediction. Through causal graphs, we were able to identify direct interactions between omics data of LUAD, furthering the potential clinical utility of multimodal omics data for prognostic biomarkers.