Big Data/Machine Learning/AI
Prediction of Acute and Chronic Kidney Diseases During the Post-COVID-19 Pandemic with Machine Learning Models: Utilizing National Electronic Health Records in the US Yue Zhang* Yue Zhang Nasrollah Ghahramani Vernon M. Chinchilli Djibril M. Ba
Background: COVID-19 infections have been demonstrated to impact acute kidney injury (AKI) and chronic kidney disease (CKD). However, the application of machine learning (ML) algorithms to predict the risk of AKI and CKD during the post-pandemic period is lacking. We aimed to leverage large electronic health records (EHR) and ML algorithms to predict the risk of incident AKI and CKD in both the short and long term during the post-pandemic period and to translate our ML models into a practical webpage application.
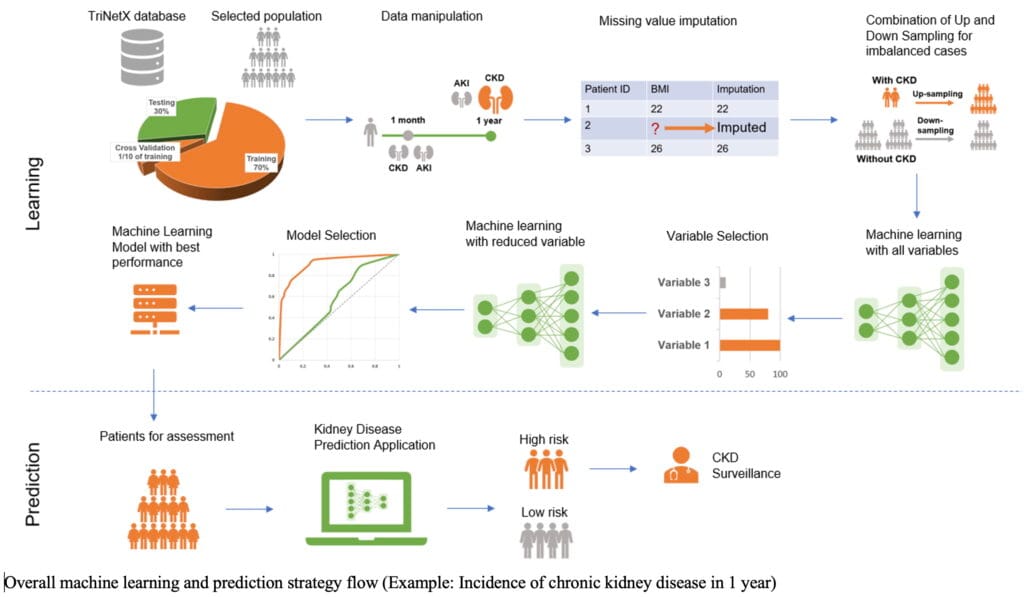
Methods: National EHR data from TriNetX were used, emulating a prospective cohort from 07/01/2022 to 03/31/2024, which was separated into training and testing datasets. A total of 69 baseline variables were included, with demographics, comorbidities, lab test results, vital signs, medication histories, hospitalization visits, and COVID-19-related variables. Two prediction windows, 1 month and 1 year from the index dates, were defined to identify the incidence of AKI and CKD. Eight machine learning models, primarily including adaptive boosting (AdaBoost), extreme gradient boosting (XGBoost), neural network (NN), and random forest (RF), were applied. Cross-validation and model tuning were conducted during the training process. Six evaluation metrics, including the area under the receiver-operating-characteristic curve (AUROC), were used to compare model performance. A combination of model-driven, data-driven, and clinical-driven methods was employed to identify the final models. An application with the final models was built using the R Shiny framework.
Results: A total of 104,565 patients were included in this study. The final models, incorporating 9 variables—primarily including eGFR, inpatient visit number, and COVID-19 infection counts—were selected. XGBoost demonstrated the best performance for predicting the incidence of AKI in 1 month (AUROC = 0.803), AKI in 1 year (AUROC = 0.799), and CKD in 1 year (AUROC = 0.894). Random Forest (RF) was selected for predicting the incidence of CKD in 1 month (AUROC = 0.896). Number of COVID-19 infections was shown to be a critical factor for inclusion in the prediction model. The final models were translated into a convenient tool to facilitate their use in clinical settings.
Conclusions: Our study demonstrates the applicability of using large national EHR data in developing high-performance machine learning models to predict AKI and CKD risks in the post-COVID-19 period. Incorporating the number of COVID-19 infections in the past year showed improved prediction performance and should be considered in future models for kidney disease prediction. A user-friendly application was created to support clinicians in risk assessment and surveillance.