Methods/Statistics
Optimal sampling strategies for observational epidemiologic studies using big data Jonathan Sterne* Jonathan Sterne Arun Karthikeyan Suseeladevi Venexia Walker Samantha Hiu Yan Ip Rochelle Knight William Whiteley Angela Wood
Background: Observational epidemiologic studies based on population-scale linked electronic health records are increasingly reported, but statistical analyses may entail substantial computational costs, or be unfeasible. Simple random sampling reduces computation time but loses precision. Subsampling based on exposure or outcome status, with analyses adjusted using inverse probability weighting, may maintain precision.
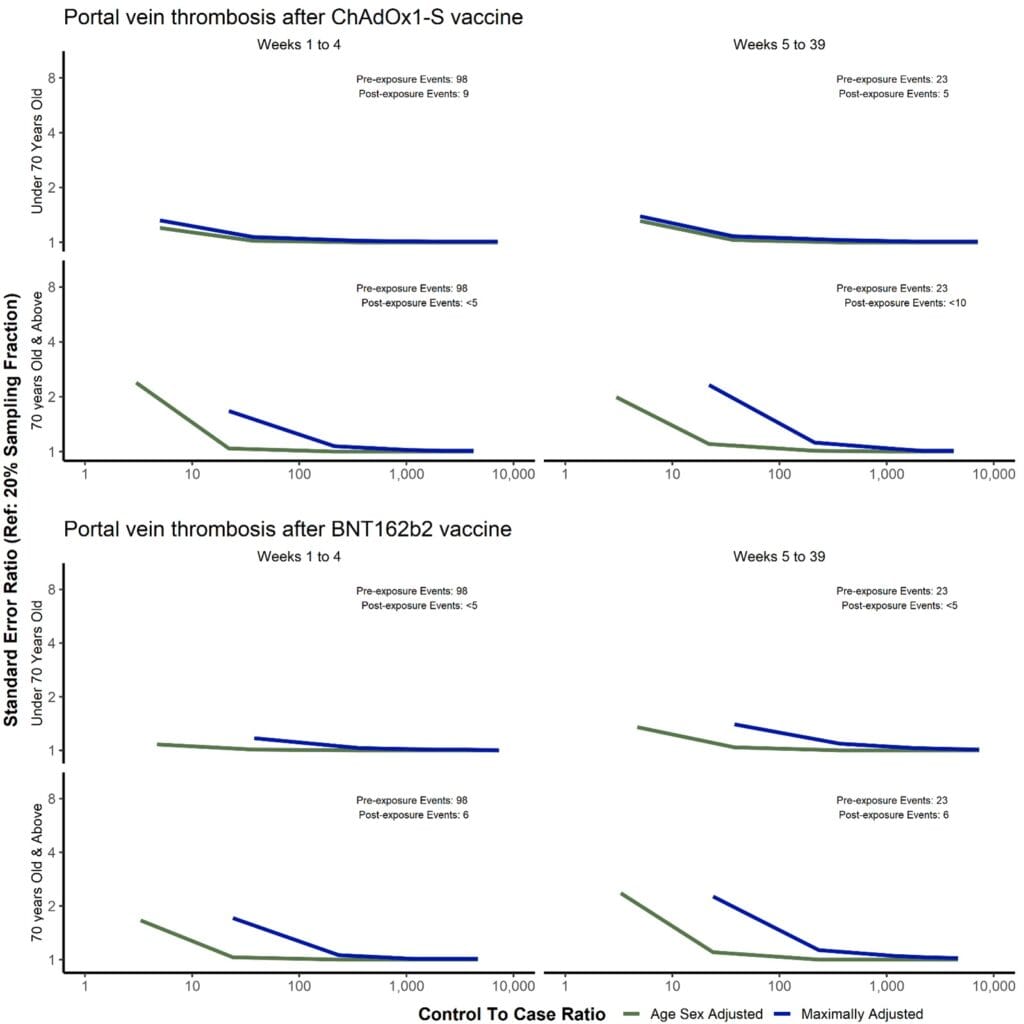
Methods: We examined the impact of different sampling fractions on precision of estimated hazard ratios (HRs) from Cox models, based on a published study (https://doi.org/10.1371/journal.pmed.1003926) using linked data on 46 million adults of associations of COVID-19 vaccination with incidence of pulmonary embolism (common outcome) and portal vein thrombosis (rare outcome). We created 50 different datasets including all people with and randomly sampled subsets of people without the outcome, for each of sampling fractions 0.001%, 0.01%, 0.1%, 1%, 2%, 5%, and 10%, and five datasets using sampling fractions 20% and 50%.
Results: Geometric mean HRs and 95% CIs were identical for sampling fractions of 10% or more. Consistent with standard case-control study theory, for the common outcome there was little or no loss of precision when there were at least 10 times as many people with versus without the outcome in the sampled dataset. However, for the rare outcome ratios of much greater than 10 were required to avoid loss of precision in multivariable models controlling for substantial numbers of confounding variables. For rare exposures, precision can be optimized by randomly sampling unexposed people without the outcome event.
Conclusions: Subsampling people based on the outcome and/or exposure, with analyses weighted to account for this, can maintain precision while reducing computation times. Such approaches to improve computational efficiency will be required as very large population scale datasets become increasingly available for epidemiologic research.