Diabetes
The molecular pathways of type 2 diabetes using proteomics, metabolic, and anthropometric profile in cross ancestry biobanks – insights on drug target discovery Junxi Liu* Junxi Liu Zhengming Chen Joanna Howson
Background
Large-scaled plasma proteomic together with genetics provides opportunities to improve understanding of type 2 diabetes (T2D) aetiology, identify biomarkers, and reveal mechanisms. The protein-level discovery is ideal for drug discovery in a precise, individual, and controlled manner.
Methods
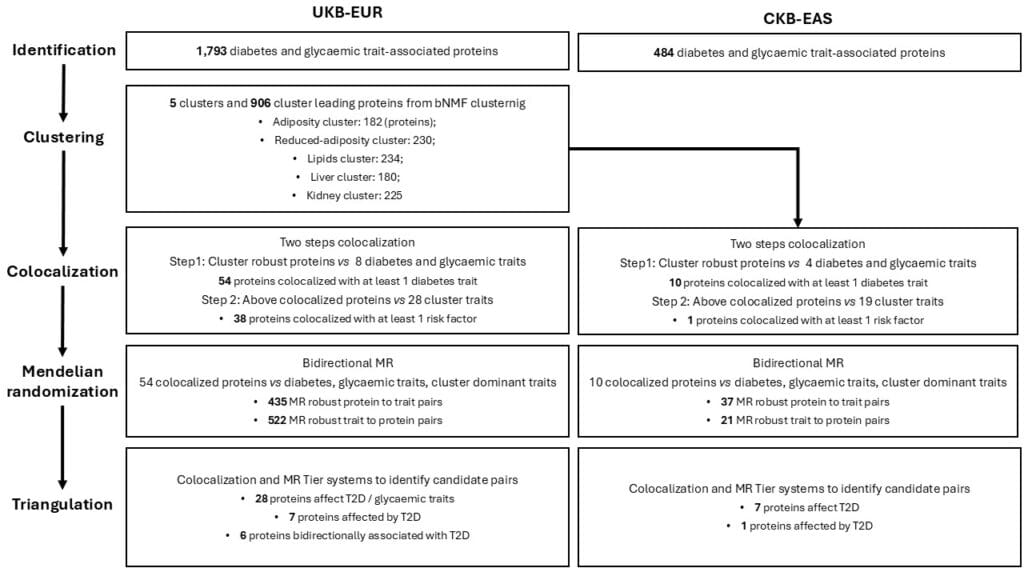
We identified proteins associated with diabetes and glycaemic traits with observational designs in UK Biobank (UKB-EUR). The Bayesian non-negative matrix factorisation (bNMF) was applied to cluster the above-obtained proteins incorporating their phenotypic associations with metabolic and anthropometric profiles. For clusters’ leading proteins, colocalization and Mendelian randomization (MR) methods were to investigate tri-variate relationships (i.e., protein – metabolic/anthropometric traits – diabetes traits). We performed the same analyses using China Kadoorie Biobank (CKB-EAS) to investigate the shared findings.
Results
1,793 proteins were observationally associated with diabetes and/or glycaemic traits. bNMF suggested five clusters (Adiposity, Reduced-adiposity, Lipids, Liver, Kidney) and 906 cluster-leading proteins (i.e., top 10% in ranking). The genetic evidence indicated effects of NCR3LG1, RTBDN, and TSPAN8 on T2D as well as T2D on FGFBP3 in both UKB-EUR and CKB-EAS. Effects of CD34, SCT, and KCTD5 on T2D were likely UKB-EUR specific; the effects of PLA2G15 and ENTR1 on T2D were likely CKB-EAS specific. We also revealed candidate proteins linking T2D via adiposity, liver, and kidney traits.
Conclusion
This study provided insights of multiple candidate proteins on T2D treatments. The protein–cluster trait associations revealed different T2D aetiology as well as linking diabetes comorbidities. There were agreements across ancestry though with heterogeneity. Further epidemiological and biological (in vitro and in vivo) validations are required.